Manuscripts under review or in revision
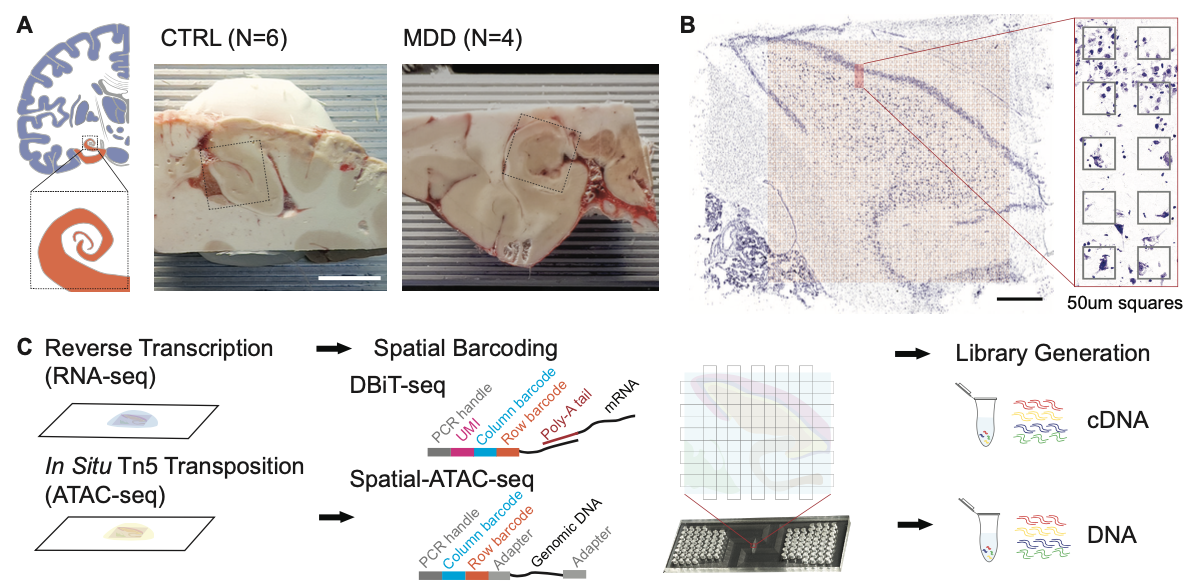
1. Xiao Y*, Su G*, Tan Y*, Asaad N, Deng Y, Li T, Lu Y, Kim D, Rosoklija GB, Enninful A, Bai Z, Liu Y, Sissoko C, Mariani M, Wu T, Nguyen P, Liang H, Santiago A, Dwork AJ, Hen R, Nolan GP, Mann JJ, Ma S, Leong KW, Boldrini M, Fan R. Spatially resolved multi-omic profiling of human hippocampus reveals region-specific alterations in major depressive disorder. (Cell, Under Review)
2. He S, Xu C, Lao HS, Chauhan S, Xiao Y, Willner MJ, Jin Y, McElroy S, Rao SB, Gogos JA, Tomer R, Azizi E, Xu B, Leong KW. Mapping morphological malformation to genetic dysfunction in blood vessel organoids with 22q11. 2 Deletion Syndrome. BioRxiv, https://doi.org/10.1101/2021.11.17.468969 (2021).
3. Nguyen PT, Tamura S, Sun E, Shi Y, Xiao Y, Lacefield C, Turi G, Hen R. Antidepressants promote developmental-like plasticity through remodeling of extracellular matrix. BioRxiv, https://doi.org/10.1101/2025.01.03.631260 (2025). (Science, Under Review)
4. Tan Y*, Kempchen TN*, Becker M, Haist M, Feyaerts D, Xiao Y, Su G, Rech AJ, Fan R, Hickey JW, Nolan GP, SPACEc: A Streamlined, Interactive Python Workflow for Multiplexed Image Processing and Analysis. BioRxiv, https://doi.org/10.1101/2024.06.29.601349 (Nature Communications, In Revision).
5. Yu L, Yang YX, Gong Z, Wan Q, Du Y, Zhou Q, Xiao Y, Zahr T, Wang Z, Yu Z, Yang K, Geng J, Fried SK, Li J, Haeusler RA, Leong KW, Bai, L, Wu Y, Sun L, Wang P, Zhu BT, Wang L, Qiang L. FcRn-dependent IgG accumulation in adipose tissue unmasks obesity pathophysiology. Cell Metabolism, Online ahead of print. DOI: 10.1016/j.cmet.2024.11.001 (2024).
23. Bai Z*, Zhang D*, Gao Y*, Tao B*, Zhang D*, Bao S*, Enninful A, Wang Y, Li H, Su G, Tian X, Zhang N, Xiao Y, Liu Y, Gerstein M, Li M#, Xing Y#, Lu J#, Xu ML#, Fan R#. Spatially Exploring RNA Biology in Archival Formalin-Fixed Paraffin-Embedded Tissue. Cell, 187(23): 6760-6779.e24 (2024).
*Technology Innovation: spatial sequencing of the non-coding RNAs
*Highlighted in Cell in the cover
22. Kim HS*, Xiao Y*, Chen X, He S, Im J, Willner MJ, Finlayson MO, Xu C, Zhu H, Choi SJ, Mosharov EV, Kim HW, Xu B, Leong KW. Chronic opioid treatment arrests neurodevelopment and alters synaptic activity in human midbrain organoids. Advanced Science, 11(21): 202400847 (2024).
21. Boldrini M, Xiao Y, Martinowich K, Singh T, Berretta S, Vallender E, Punzi G, Gürsoy G, Pantazopoulos H, Marenco S, Jabbi M, Zhu C, Hyde T, Zody M, Lewis D, Turecki G, Arango V, Pevsner J, Roussos P, Lehner T, Mann JJ. Omics approaches to investigate the pathogenesis of suicide. Biological Psychiatry, 96(15): 919-928 (2024).
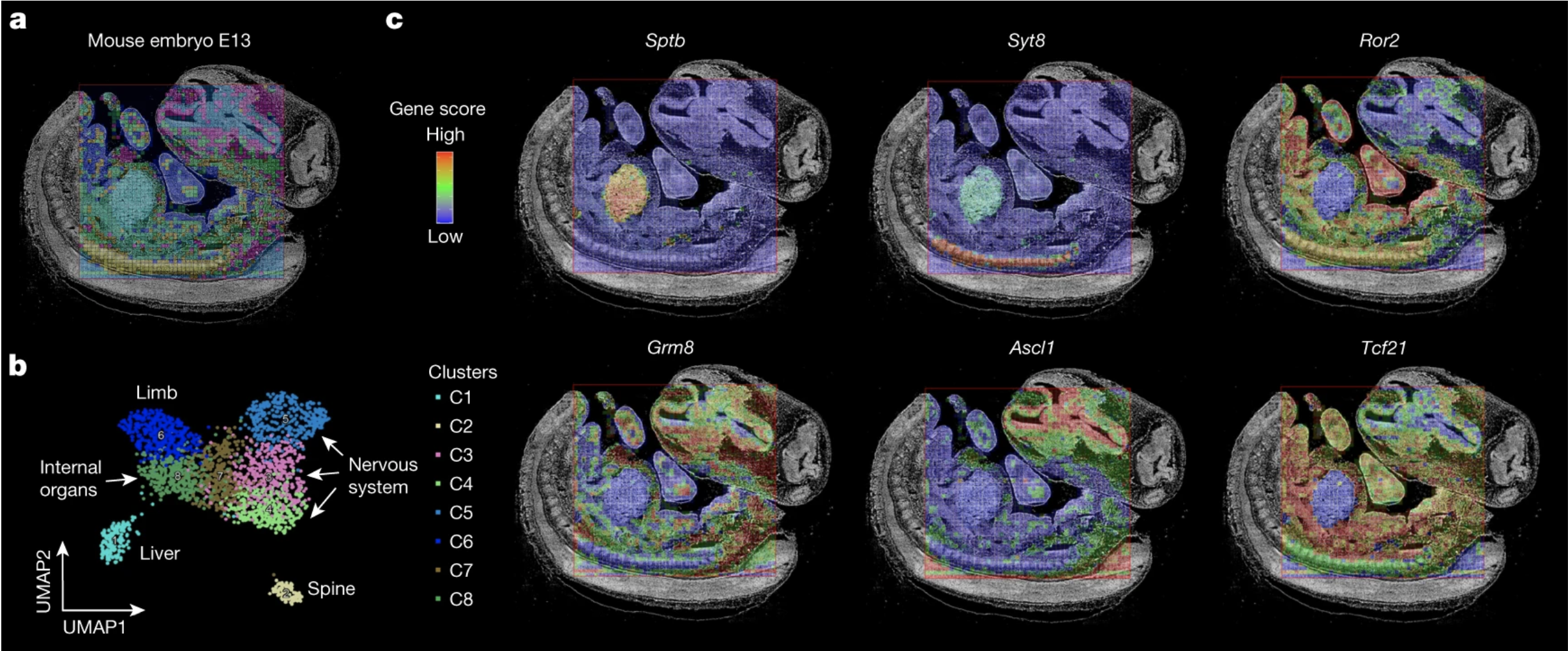
20. Zhang D*, Deng Y*, Kukanja P, Kukanja P, Agirre E, Bartosovic M, Dong M, Ma C, Ma S, Su G, Bao S, Liu Y, Xiao Y, Ma S, Rosoklija GB, Dwork AJ, Mann JJ, Leong KW, Boldrini M, Wang L, Haeussler M, Raphael BJ, Kluger Y, Castelo-Branco G, Fan R. Spatial epigenome–transcriptome co-profiling of mammalian tissues. Nature, 616(7955): 113–122 (2023).
*Technology innovation: Spatial co-sequencing of mRNA and open chromatin platform
*Highlighted in Nature Research Briefing
Gene expression and epigenetic regulation co-mapped in brain tissues. Nature, Advance online publication. https://www.nature.com/articles/d41586-023-00436-z (2023).
*Highlighted in Nature Reviews Genetics Research Highlight
Layering epigenomic and transcriptomic space. Nature Reviews Genetics, Advance online publication. https://www.nature.com/articles/s41576-023-00596-8 (2023).
19. Chen AT*, Xiao Y*, Tang X*, Baqri M, Gao X, Reschke M, Zhou Y, Deng G, Zhang S, Deng Y, Bai Z, Kim D, Huttner A, Kunes R, Saltzman WM, Fan R, Zhou J. Cross-platform analysis reveals cellular and molecular landscape of glioblastoma invasion. Neuro-Oncology, 25(3): 482-494 (2023).
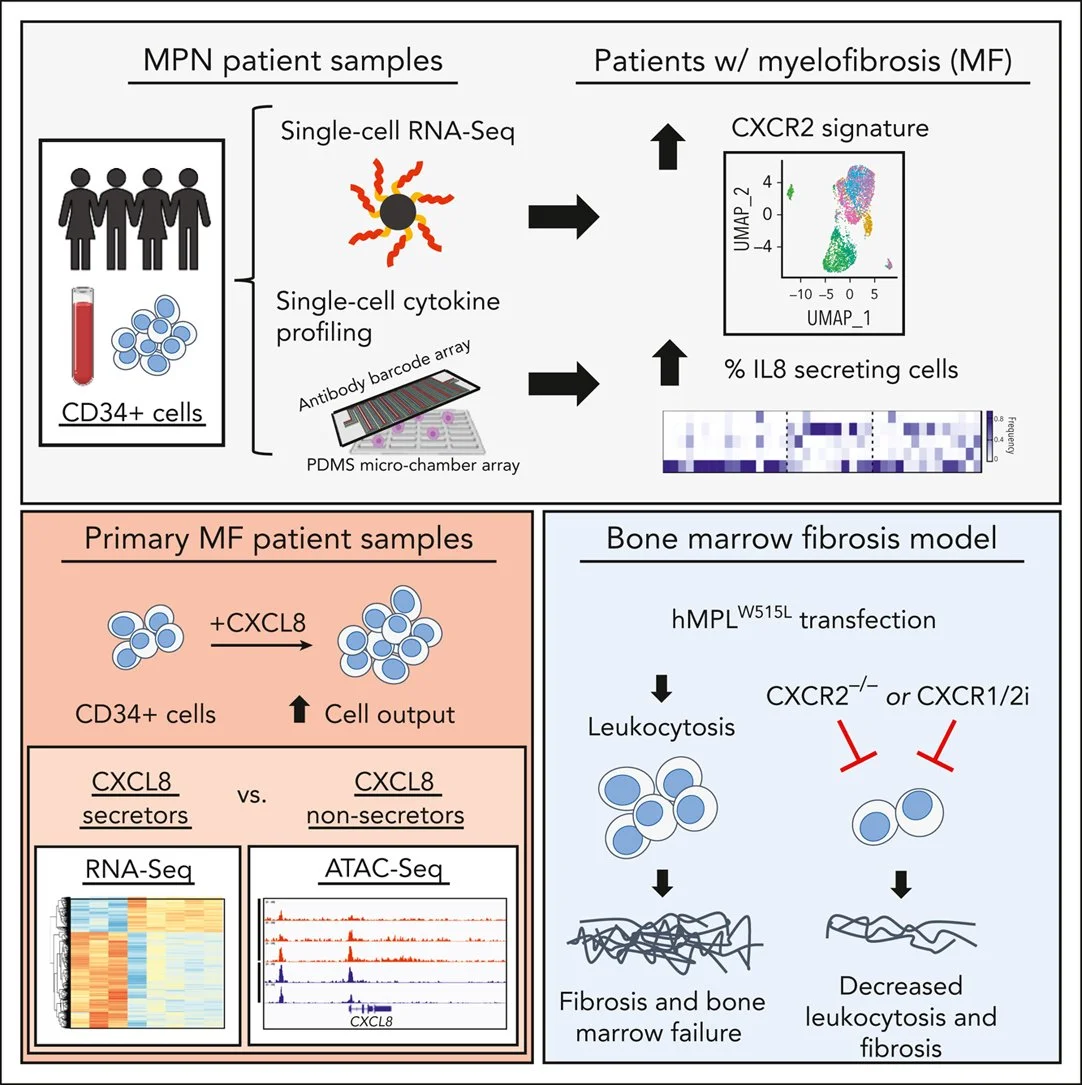
18. Dunbar A, Kim D, Lu M, Farina M, Bowman RL, Yang JL, Park YC, Karzai A, Xiao W, Zaroogian Z, O'Connor K, Mowla S, Gobbo F, Verachi P, Martelli F, Sarli G, Xia L, Elmansy N, Kleppe M, Chen Z, Xiao Y, McGovern EM, Snyder J, Krishnan A, Hill CE, Cordner KB, Zouak A, Salama ME, Yohai J, Tucker E, Chen JJ, Zhou J, McConnell TS, Migliaccio AR, Koche RP, Rampal RK, Fan R, Levine R, Hoffman R. CXCL8/CXCR2 signaling mediates bone marrow fibrosis and represents a therapeutic target in myelofibrosis. Blood, 141 (20): 2508–2519 (2023).
*Highlighted in Blood Editorial Commentary
Shelter from the cytokine storm in myelofibrosis. Blood, 141 (20): 2415–2416 (2023). https://www.sciencedirect.com/science/article/pii/S0006497123005633
17. Wan Q*, Huang B*, Li T, Xiao Y, He Y, Du W, Wang BZ, Dakin GF, Rosenbaum M, Goncalves MD, Chen S, Leong KW, Qiang L. Selective targeting of visceral adiposity by polycation nanomedicine. Nature Nanotechnology, 17(12):1311-1321 (2022).
*Highlighted in Nature Nanotechnology Research Briefing
Targeting and reducing abdominal fat using polycations. Nature Nanotechnology 17(12):1249-1250 (2022).
16. Fang F, Xiao Y, Zelzer E, Leong KW, Thomopoulos S. A mineralizing pool of Gli1-expressing progenitors builds the tendon enthesis and demonstrates therapeutic potential. Cell Stem Cell, 29(12): 1669-1684 (2022).
*Highlighted in Cell Stem Cell Editorial Preview
Craft A and Galloway J. Specialized cells for building tissue bridges. Cell Stem Cell, 29(12):1615-1616 (2022).
15. Deng Y, Bartosovic M, Ma S, Zhang D, Kukanja P, Xiao Y, Su G, Liu Y, Qin X, Rosoklija GB, Dwork AJ, Mann JJ, Xu ML, Halene S, Craft JE, Leong KW, Boldrini M, Castelo-Branco G, Fan R. Spatial profiling of chromatin accessibility in mouse and human tissues. Nature, 609(7926):375-383 (2022).
*Technology innovation: Spatial ATAC-seq Platform for Detection of Open Chromatin
*Featured in Nature Structural & Molecular Biology Research Highlight
Chromatin found in space, Perdigoto C. Nature Structural & Molecular Biology, 29 (843) (2022).
14. Bai Z, Woodhouse S, Kim D, Lundh S, Sun H, Deng Y, Xiao Y, Barrett DM, Myers RM, Grupp SA, June CH, Melenhorst JJ, Camara PG, Fan R. Single-cell antigen-specific activation landscape of CD19 CAR T cell infusion product predicts clinical response and relapse in patients with acute lymphoblastic leukemia. Science Advances, 8(23): abj2820 (2022).
13. Xiao Y, Wang Z, Zhao M, Deng Y, Yang M, Su G, Yang K, Qian C, Hu X, Liu Y, Geng L, Xiao Y, Zou Y, Tang X, Liu H, Xiao H and Fan R. Single-Cell Transcriptomics Revealed Subtype-Specific Tumor Immune Microenvironments in Human Glioblastomas. Frontiers in Immunology, 13:914236 (2022).
12. Liu J*, Wang X*, Chen AT*, Gao X*, Himes BT, Zhang H, Chen Z, Wang J, Sheu WC, Deng G, Xiao Y, Zou P, Zhang S, Liu F, Zhu Y, Fan R, Patel TR, Saltzman WM, Zhou J. ZNF117 regulates glioblastoma stem cell differentiation towards oligodendroglial lineage. Nature Communications, 13:2196 (2022).
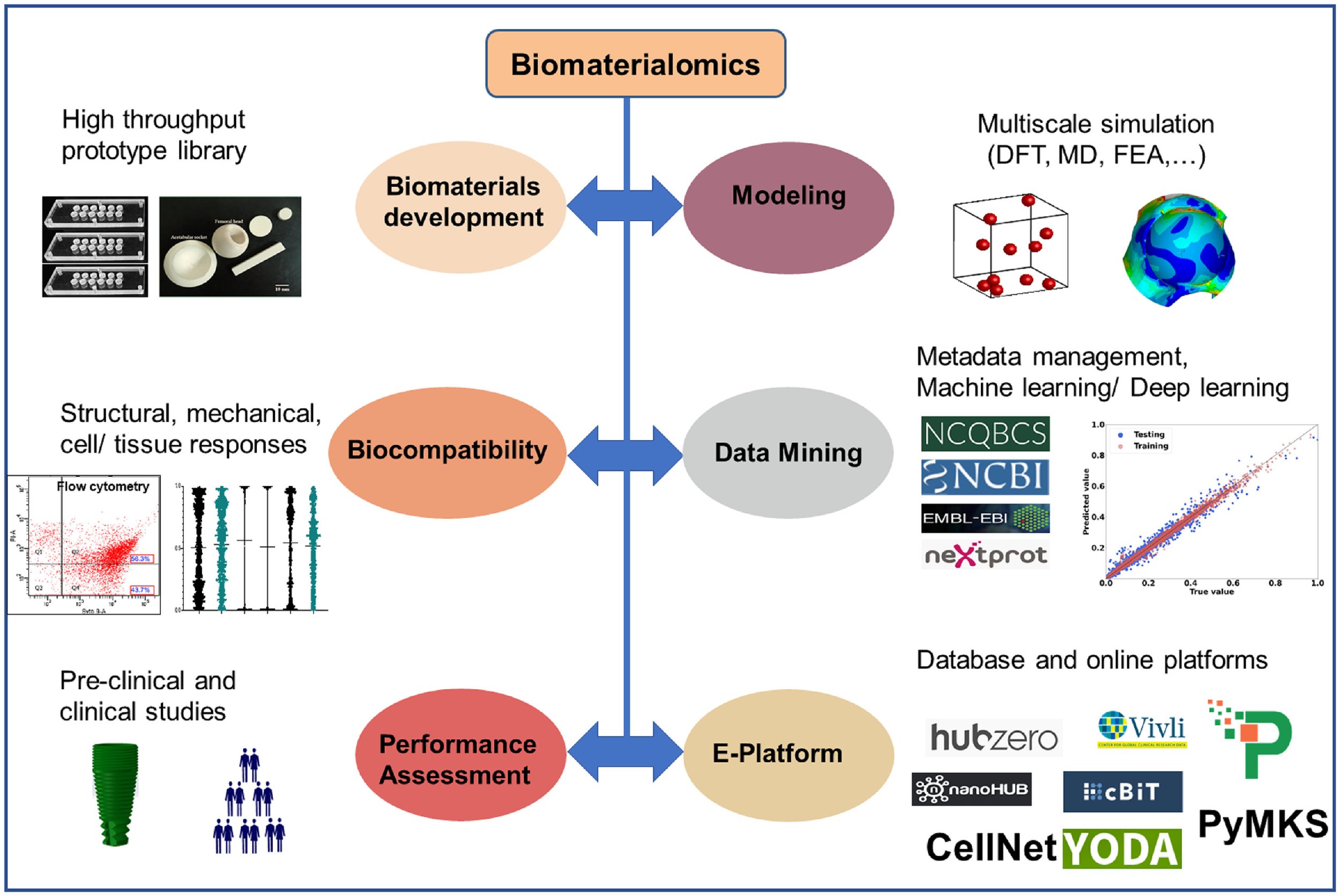
11. Basu B, Gowtham, NH, Xiao Y, Kalidindi SR, Leong KW. Biomaterialomics: Data-Driven Pathways to Next-Generation Biomaterials. Acta Biomaterialia, 143: 1-25 (2022).
10. Willner MJ*, Xiao Y*, Kim HS, Chen XJ, Xu B, Leong KW. Modeling SARS-CoV-2 infection in individuals with opioid usage disorder with brain organoids. Journal of Tissue Engineering, 12: 2041731420985299 (2021).
9. Liu Y, Yang M, Deng Y, Su G, Enniful A, Guo C, Tebaldi T, Zhang D, Kim D, Bai Z, Norris E, Pan A, Li J, Xiao Y, Halene S, Fan R. High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue. Cell, 183(6): 1665-1681.e18 (2020).
*Technology innovation: Spatial mRNA-seq Platform
8. Huang D, Gibeley SB, Xu C, Xiao Y, Celik O, Ginsberg H, Leong KW. Engineering liver microtissues for disease modeling and regenerative medicine. Advanced Functional Materials, 30(44): 201909553 (2020).
7. Bai Z, Deng Y, Kim D, Chen Z, Xiao Y, Fan R. An Integrated Dielectrophoresis-Trapping and Nanowell Transfer Approach to Enable Double-Sub-Poisson Single-Cell RNA Sequencing. ACS Nano, 14(6): 7412-7424 (2020).
6. Xiao Y, Kim D, Dura B, Zhang K, Yan R, Li H, Han E, Ip J, Zou P, Liu J, Chen AT, Vortmeyer AO, Zhou J, Fan R. Ex vivo dynamics of human glioblastoma cells in a microvasculature-on-a-chip system correlates with tumor heterogeneity and subtypes. Advanced Science, 6(8): 1801531 (2019).
*Selected as Inside Front Cover
5. Xiao Y, Liu C, Chen Z, Blatchley M, Gerecht S, Fan R. Senescent fibroblasts promote microvasculature formation in vitro. Advanced Biosystems, 3(8): 1900089 (2019).
*Selected as Inside Front Cover
4. Chen Z, Lu Y, Zhang K, Xiao Y, Lu J, Fan R. Multiplexed, sequential secretion analysis of the same single cells reveals distinct effector response dynamics dependent on the initial basal state. Advanced Science, 6(9): 1801631 (2019).
*Selected as Inside Back Cover
3. Xhangolli I, Dura B, Lee G, Kim D, Xiao Y, Fan R, Single-cell integrative analyses of CAR-T cell activation reveals a predominantly TH1/TH2 mixed response independent of differentiation, Genomics, Proteomics & Bioinformatics, 17(2): 129-139 (2019).
*Selected as Front Cover
2. Sivarapatna A, Ghaedi M, Xiao Y, Han E, Aryal B, Zhou J, Fernandez-Hernando C, Qyang Y, Hirschi KK, Niklason LE, Engineered Microvasculature in PDMS Networks Using Endothelial Cells Derived from Human Induced Pluripotent Stem Cells. Cell Transplantation, 26(8): 1365-1379 (2017).
1. Campos M, Buchanan A, Yu F, Barbalic M, Xiao Y, Chambless LE, Wu KK, Folsom AR, Boerwinkle E, Dong JF, Influence of single nucleotide polymorphisms in factor VIII and von Willebrand factor genes on plasma factor VIII activity: the ARIC Study. Blood, 119(8): 1929-1934 (2012).